Our research
Proteins are the building blocks of living things, miniature motors that make all of your cells function. Proteins embedded in cell membranes filter out toxic materials or uptake necessary nutrients. Meanwhile, malfunctioning proteins are responsible for a slew of disorders, including Alzheimer’s, type II diabetes and Parkinson’s. Our group works on understanding and designing small molecules and peptides for therapeutic applications such as finding new analgesics for treatment of chronic pain disorders or correcting dysregulation of proteins that lead to Alzheimer’s.
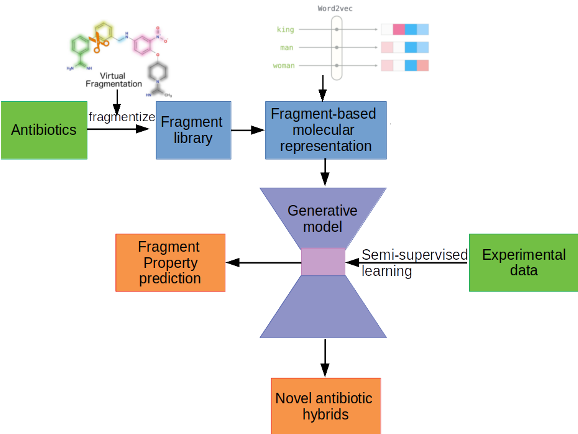
Hybrid antibiotic design through interpretable deep learning
With dwindling drug leads and increasing fatalities from multidrug-resistant bacteria, we are teetering on the brink of the post-antibiotic era. New design techniques and approaches for both understanding of antibiotic resistance and design of novel therapeutics are urgently needed. Recently, there has been promising experimental work in the field of antibiotic hybrids, in which sets of different pharmacophores are linked in novel ways, which may lead to molecules with enhanced efficacy or even entirely novel properties.
We are working on exploring the use of a fragment-based approach for novel antibiotic hybrid design through generative deep learning, in which a library of fragments relevant to antibiotic applications is used as a basis for a generative model that produces novel antibiotic hybrids through linking, growth and merging, with a particular emphasis on interpretability as well as candidate generation.
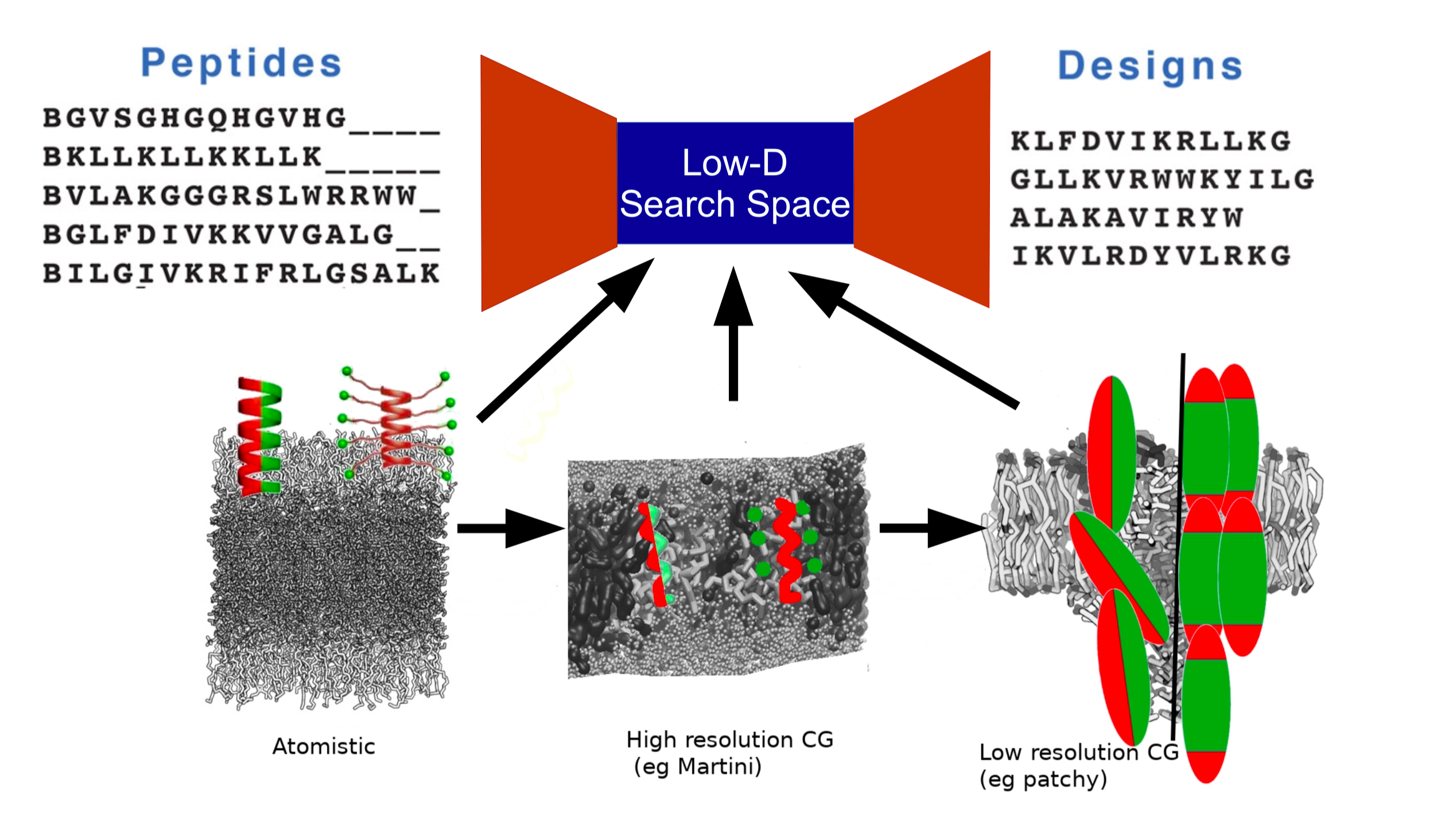
Antimicrobial Peptide Design
Although design of novel antibiotics is one approach to combat the growing problem of multidrug resistance, it is unlikely to be sufficient on its own and a multipronged approach is warranted. Another promising line of research for novel drugs with antimicrobial action is the design of peptides with antimicrobial properties that preferentially disrupt the membranes of bacterial cells while displaying a lower affinity for mammalian cells. This is still an active area of research, with only a few candidate peptides having reached clinical trials, due to the continued existence of unwanted side effects such as serum binding or the requirement of potentially-toxic concentrations to be effective.
We are working on designing a deep learning model informed by multiscaled molecular dynamics, wherein generative learning is used to iteratively create potential AMP candidates that are assessed on multiple scales using molecular dynamics, from high-resolution atomistic molecular dynamics to assess detailed properties on short time scales, to low resolution “spherical peptide in a vacuum” models to assess penetration of candidates into the cell membrane.
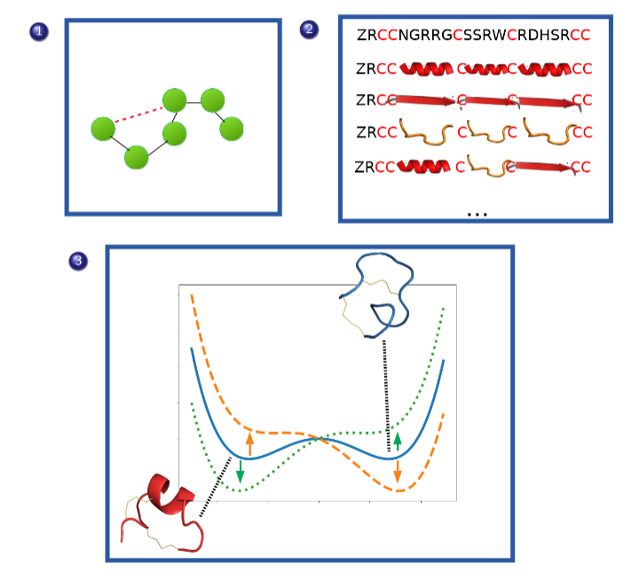
Theory of disulfide bonds for toxin-based therapeutics
Owing to their high specificity and binding affinity for various receptors involved in different biological pathways, toxins have for a long time been considered a rich natural source of therapeutic leads. A large and intriguing class of toxins is those that are short, cysteine-rich peptides whose structures are largely controlled by the multiple disulfide bonds that form between the cysteines. Historically, one difficulty in assessing the structure and hence the proposed mechanisms of such toxins has been that it can prove difficult to controllably fold them to their native stable states in vitro. Indeed, there is evidence that certain toxins exist even in vivo in metastable states that are nonetheless potent and in certain cases the “non-native” fold demonstrates higher selectivity and binding affinity which leads to the question of whether we can control these metastable states through sequence, environment or kinetic engineering.
We are exploring ways to bring together constrained polymer theories with bond-breaking force field models to map out the free energy landscapes of disulfide-rich toxins.